BLAST/BLAT RESULTS |
||
|
|

BLASTN 2.2.17 [Aug-26-2007]
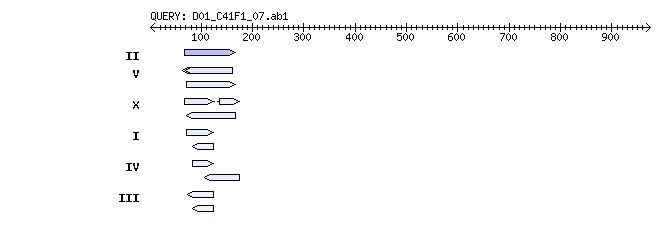
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= D01_C41F1_07.ab1 ABIX Testing -- no comment (974 letters) Database: c_elegans nucleotide release [WS195] 7 sequences; 100,281,427 total letters Searching..................................................done Score E Sequences producing significant alignments: (bits) Value II 198 1e-49  [Alignment][Genome View] [Alignment][Genome View]V 80 9e-14  [Alignment][Genome View] [Alignment][Genome View]X 78 4e-13  [Alignment][Genome View] [Alignment][Genome View]I 64 5e-09  [Alignment][Genome View] [Alignment][Genome View]IV 58 3e-07  [Alignment][Genome View] [Alignment][Genome View]III 44 0.005  [Alignment][Genome View] [Alignment][Genome View]>II Length = 15279324  [Genome View] [Genome View]Score = 198 bits (100), Expect = 1e-49 Identities = 100/100 (100%) Strand = Plus / Plus Query: 67 tacacgagtcacatcagagaaaagccaataatcactagaagcaatgtcaggagaatttgg 126 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| Sbjct: 2284481 tacacgagtcacatcagagaaaagccaataatcactagaagcaatgtcaggagaatttgg 2284540 Query: 127 ttgctgatagaatcatccatccatattttgccagctcctg 166 |||||||||||||||||||||||||||||||||||||||| Sbjct: 2284541 ttgctgatagaatcatccatccatattttgccagctcctg 2284580 >V Length = 20919569  [Genome View] [Genome View]Score = 79.8 bits (40), Expect = 9e-14 Identities = 85/99 (85%), Gaps = 7/99 (7%) Strand = Plus / Minus Query: 69 cacgagtcacatcagagaaaagccaataatcactagaagca-----atgtcaggagaatt 123 ||||||||| ||||||| |||||||||||||||| | |||| ||||||| |||||| Sbjct: 670179 cacgagtcagatcagagtaaagccaataatcacttggagcacttggatgtcagaagaatt 670120 Query: 124 tggttgctga--tagaatcatccatccatattttgccag 160 ||| ||||| ||||||||||||||||||||||||||| Sbjct: 670119 cggtggctgagatagaatcatccatccatattttgccag 670081 Score = 67.9 bits (34), Expect = 3e-10 Identities = 82/97 (84%), Gaps = 2/97 (2%) Strand = Plus / Plus Query: 72 gagtcacatcagagaaaagccaataatcactagaagcaatgtcaggagaatttggttgct 131 |||||| ||| |||||||||||||||||||| ||||||||||||| |||||||| | Sbjct: 10308937 gagtcagatcggagaaaagccaataatcacttagagcaatgtcaggaaaatttggtggta 10308996 Query: 132 ga--tagaatcatccatccatattttgccagctcctg 166 || ||||||| |||||||||||||| ||| ||||| Sbjct: 10308997 gagggagaatcacccatccatattttgtcagttcctg 10309033 Score = 61.9 bits (31), Expect = 2e-08 Identities = 52/59 (88%) Strand = Plus / Minus Query: 64 gattacacgagtcacatcagagaaaagccaataatcactagaagcaatgtcaggagaat 122 |||| ||| ||||| ||| |||| ||||||||||||||| |||| |||||||||||||| Sbjct: 11591415 gattgcacaagtcagatcggagagaagccaataatcacttgaagaaatgtcaggagaat 11591357 >X Length = 17718854  [Genome View] [Genome View]Score = 77.8 bits (39), Expect = 4e-13 Identities = 51/55 (92%) Strand = Plus / Plus Query: 68 acacgagtcacatcagagaaaagccaataatcactagaagcaatgtcaggagaat 122 |||||||||| |||||||||||||||||||||||| | || |||||||||||||| Sbjct: 6467872 acacgagtcagatcagagaaaagccaataatcacttggagaaatgtcaggagaat 6467926 Score = 60.0 bits (30), Expect = 9e-08 Identities = 81/97 (83%), Gaps = 2/97 (2%) Strand = Plus / Minus Query: 72 gagtcacatcagagaaaagccaataatcactagaagcaatgtcaggagaatttggttgct 131 |||||| ||| |||||||||||||||||||| ||||||||||||| |||| ||| | Sbjct: 6418606 gagtcagatcggagaaaagccaataatcacttagagcaatgtcaggaaaattcggtggta 6418547 Query: 132 ga--tagaatcatccatccatattttgccagctcctg 166 || ||||||| |||||||||||||| ||| ||||| Sbjct: 6418546 gagggagaatcacccatccatattttgtcagatcctg 6418510 Score = 46.1 bits (23), Expect = 0.001 Identities = 35/39 (89%) Strand = Plus / Plus Query: 135 agaatcatccatccatattttgccagctcctgaatcact 173 ||||| ||||||||||||||| |||| ||||| |||||| Sbjct: 339575 agaattatccatccatattttaccagttcctgcatcact 339613 >I Length = 15072421  [Genome View] [Genome View]Score = 63.9 bits (32), Expect = 5e-09 Identities = 47/52 (90%) Strand = Plus / Plus Query: 72 gagtcacatcagagaaaagccaataatcactagaagcaatgtcaggagaatt 123 |||||| ||| |||||||||||||||||||| |||||||||||||||||| Sbjct: 11940540 gagtcagatcggagaaaagccaataatcacttagagcaatgtcaggagaatt 11940591 Score = 40.1 bits (20), Expect = 0.079 Identities = 35/40 (87%) Strand = Plus / Minus Query: 83 gagaaaagccaataatcactagaagcaatgtcaggagaat 122 |||| ||||||| ||||||| | || |||||||||||||| Sbjct: 4099100 gagagaagccaacaatcacttggagaaatgtcaggagaat 4099061 >IV Length = 17493784  [Genome View] [Genome View]Score = 58.0 bits (29), Expect = 3e-07 Identities = 38/41 (92%) Strand = Plus / Plus Query: 83 gagaaaagccaataatcactagaagcaatgtcaggagaatt 123 |||||||||||||||||||| |||||||||||||||||| Sbjct: 3462187 gagaaaagccaataatcacttagagcaatgtcaggagaatt 3462227 Score = 54.0 bits (27), Expect = 5e-06 Identities = 60/70 (85%), Gaps = 2/70 (2%) Strand = Plus / Minus Query: 106 agcaatgtcaggagaatttggttgctgatagaat--catccatccatattttgccagctc 163 |||||||||| ||||||| ||| | ||| |||| |||||| ||||||||||||||||| Sbjct: 17146338 agcaatgtcaagagaattcggtggttgagagaaaaccatccacccatattttgccagctc 17146279 Query: 164 ctgaatcact 173 ||| |||||| Sbjct: 17146278 ctgcatcact 17146269 >III Length = 13783681  [Genome View] [Genome View]Score = 44.1 bits (22), Expect = 0.005 Identities = 43/50 (86%) Strand = Plus / Minus Query: 73 agtcacatcagagaaaagccaataatcactagaagcaatgtcaggagaat 122 ||||| ||| |||| ||||||||||||||| | || |||| ||||||||| Sbjct: 7478193 agtcagatcggagagaagccaataatcacttggagaaatgccaggagaat 7478144 Score = 40.1 bits (20), Expect = 0.079 Identities = 35/40 (87%) Strand = Plus / Minus Query: 83 gagaaaagccaataatcactagaagcaatgtcaggagaat 122 |||| ||||||| ||||||| | || |||||||||||||| Sbjct: 5098492 gagagaagccaacaatcacttggagaaatgtcaggagaat 5098453 |
|